Introduction
Some time ago I wrote about the support for R users within Dataiku. The software can really boost the productivity of R users by having:
- Managed R code environments,
- Support for publishing of R markdown files and shiny apps in Dataiku dashboards,
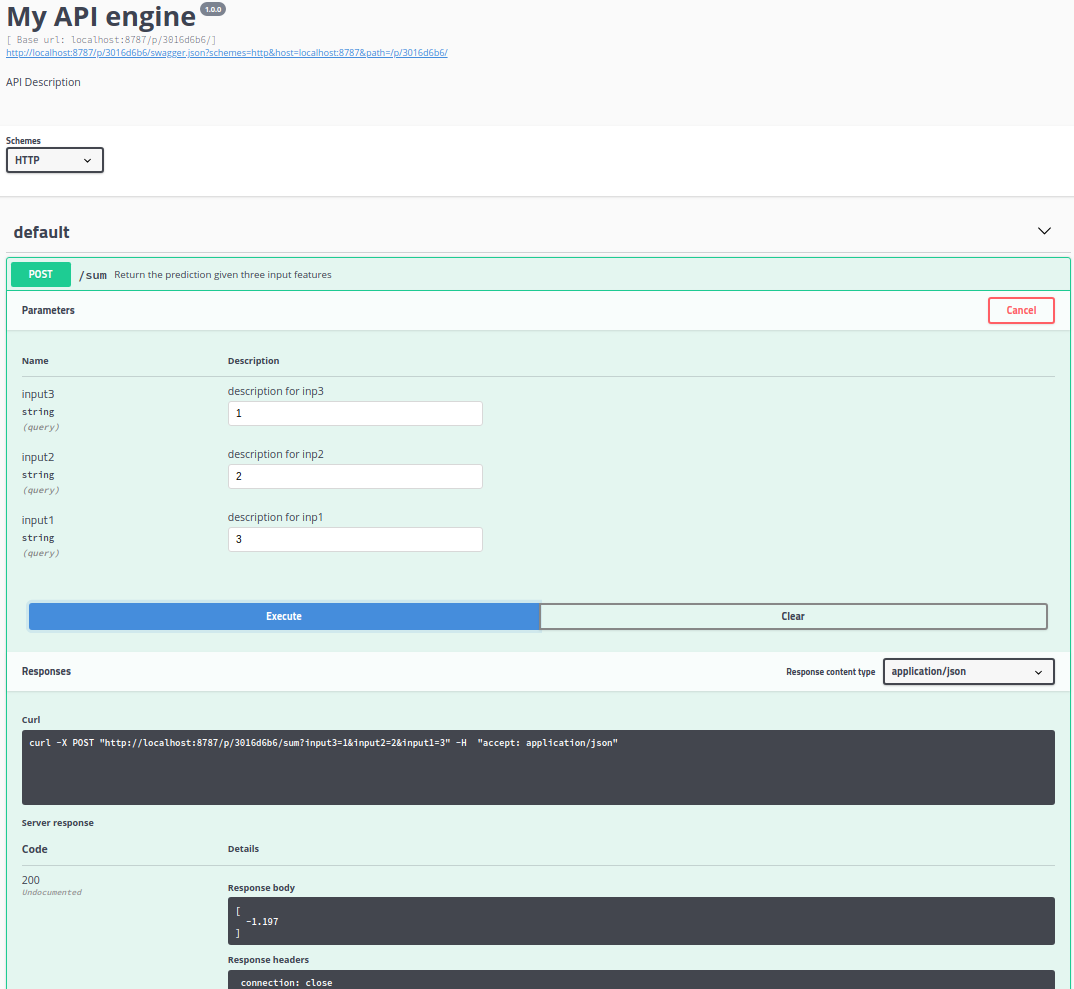
- The ability to easily create and deploy API’s based on R functions.
See the blog post here.
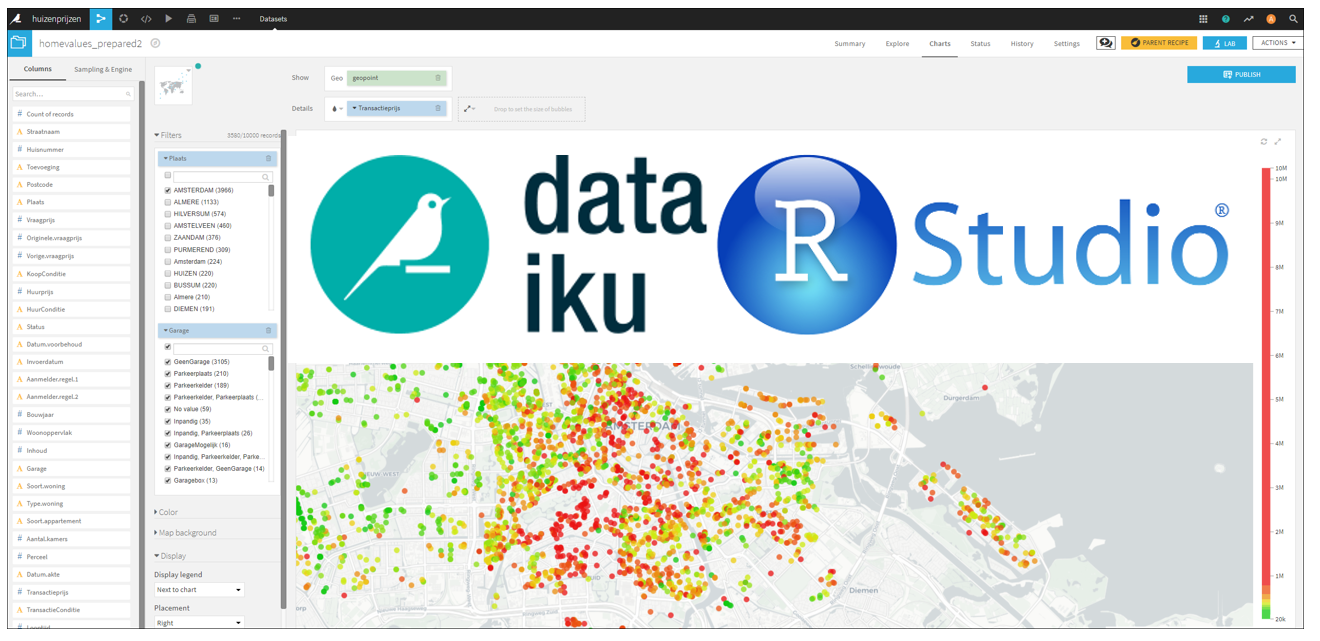
With the 5.1 release of Dataiku two of my favorite data science tools are brought closely together :-). Within Dataiku there is now a nice integration with RStudio. And besides this, there is much more new functionality in the system, see the 5.1 release notes.
RStudio integration
From time to time, the visual recipes to perform certain tasks in Dataiku are not enough for what you want to do. In that case you can make use of code recipes. Different languages are supported, Python, Scala, SQL, shell and R. To edit the R code you are provided with an editor in Dataiku, it is however, not the best editor I’ve seen. Especially when you are used to the RStudio IDE 🙂
There is no need to create a new R editor, that would be a waist of time and resources. In Dataiku you can now easily use the RStudio environment in your Dataiku workflow.

Suppose you are using an R code recipe in your workflow, then in the code menu in Dataiku you can select ‘RStudio server’. It will bring you to an RStudio session (embedded) in the Dataiku interface (either on the same Dataiku server or on a different server).
In your (embedded) RStudio session you can easily ‘communicate’ with the Dataiku environment. There is a ‘dataiku’ R package that exposes functions and RStudio add-ins to make it easy to communicate with Dataiku. You can use it to bring over R code recipes and / or data from Dataiku. This will allow you to edit and test the R code recipe in RStudio with all the features that a modern IDE should have.

Once you are done editing and testing in RStudio you can then upload the R code recipe back to the Dataiku workflow. If your happy with the complete Dataiku workflow, put it into production, i.e. run it in database, or deploy it as an API. See this short video for a demo.
Stay tuned for more Dataiku and R updates….. Cheers, Longhow.